
|
Macromolecular Networks Underlying Chloroplast Biogenesis This project is funded by National Science Foundation grant IOS-0922560 |

|
|
|
||
|
Home
Personnel Publications Contact Us Resources: Protocols Sequence-indexed Mu Insertions PML Mutant Collection POGs Database Plant Proteome Database Outreach Programs: Barkan Lab High School Mentorship Program Tutorials for School Kids Barkan Lab Website van Wijk Lab Website |
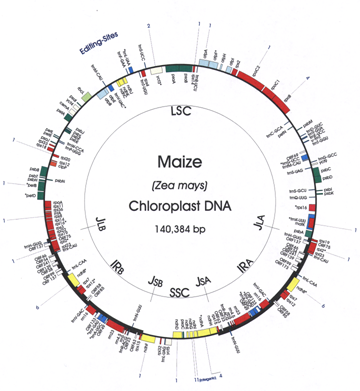
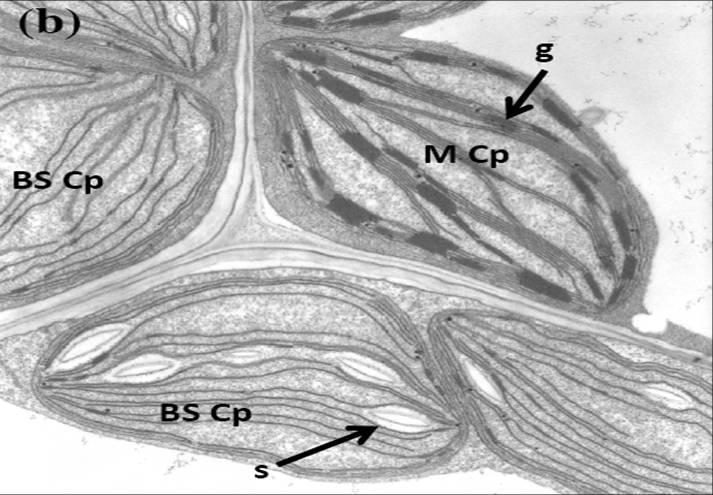
Alice Barkan, PI, University of Oregon Klaas van Wijk, coPI, Cornell University Chloroplasts are a defining feature of plants, with diverse metabolic functions that profoundly impact plant yield and response to environmental stress. Photosynthesis is the primary raison d’etre of the chloroplast, and involves a complex machinery whose biogenesis requires a series of functionally and physically-connected gene expression and assembly processes. The goal of this project is to advance understanding of the network of DNA/RNA/protein interactions that underlie the biogenesis of the photosynthetic apparatus in chloroplasts. Macromolecular assemblies selected for study are anticipated to be rich in proteins of unknown function, and are being dissected through concerted genetic, proteomic, and ribonomic analyses. Maize is our primary experimental organism because its attributes make it especially well suited for these methods, it is an important crop species, and it is a prime model for understanding C4 photosynthesis. (1) Nuclear genes required for chloroplast biogenesis will be discovered through a high-throughput forward-genetic strategy that combines “Next Generation” sequencing with a deep collection of transposon-induced non-photosynthetic maize mutants. Phenotypic data that place mutants into under-studied functional classes are being used to select 100 lines for this analysis. (2) Protein components of immunopurified macromolecular assemblies will be identified through high sensitivity mass spectrometry. Antibody targets will include: nascent peptide chains associated with translating polysomes to immunopurify “factories” for the synthesis and assembly of chloroplast-encoded proteins; proteins associated with the chloroplast chromosome; and proteins at the interface of the chloroplast gene expression and protein folding/assembly machineries. Newly-identified proteins will be functionally characterized by reverse-genetics and phenotypic analyses, and by identification of their nucleic acid ligands (where relevant). (3) Genome-wide RNA/DNA coimmunoprecipitation assays will be used to identify the nucleic acids associated with nucleic-acid interacting proteins identified via the genetic and proteome approaches, and to explore the functions of organelle-dedicated RNA binding protein families. |
|